
Reference map of the extracellular proteome of P. chrysogenum Wisconsin 54-1255

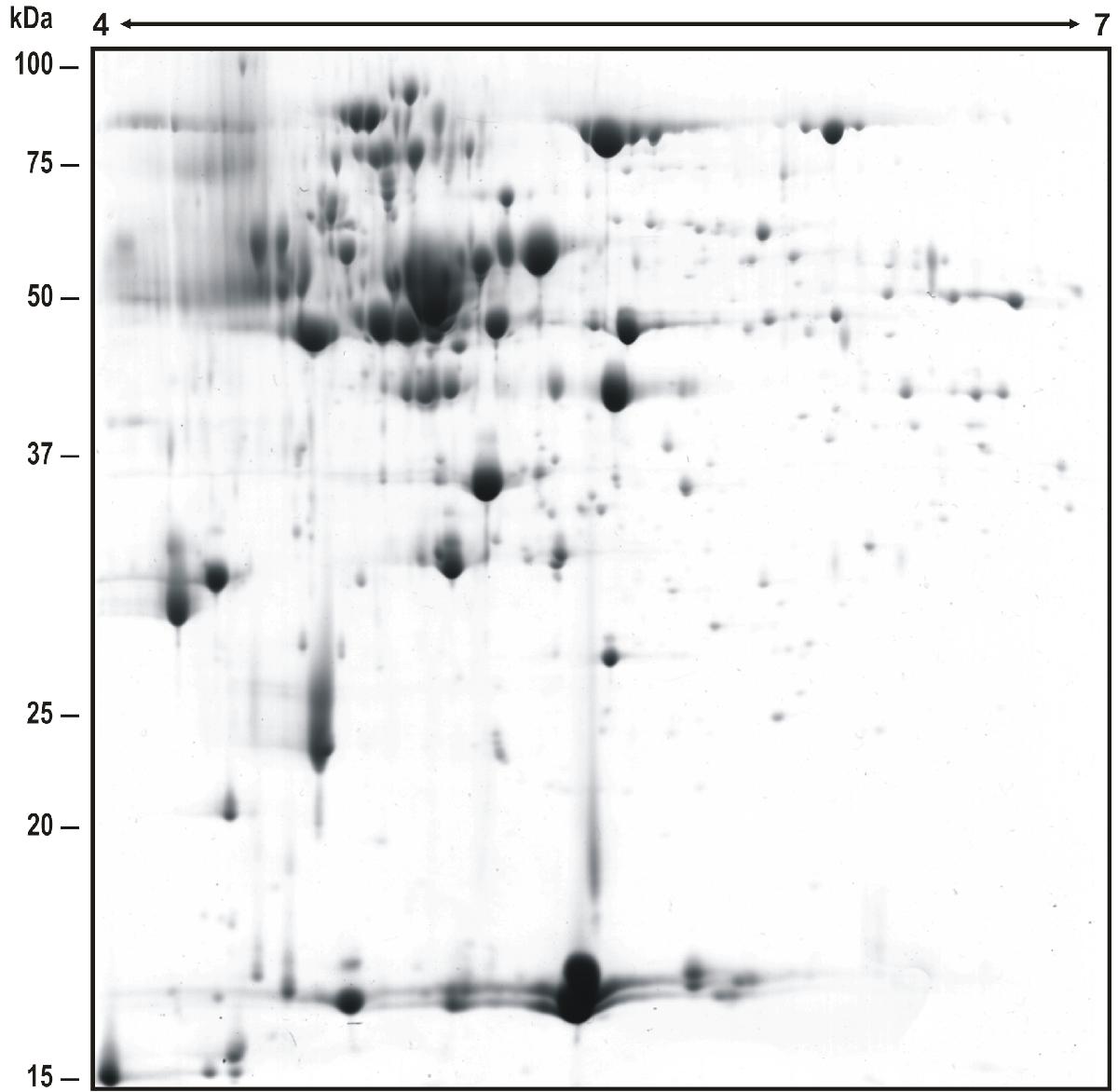
Reference map of the extracellular proteome of Penicillium chrysogenum. Proteins were separated by 2-DE using 18-cm wide-range IPG strips (pH 4-7 NL) and 12.5% SDS-PAGE gel. "Blue Silver" Colloidal Coomassie was used as staining method. A total amount of 279 spots were identified by PMF and tandem mass spectrometry. Molecular weights are shown on the left. Information of the identified proteins is shown when the mouse pointer is moved over a spot: Spot number (correlated with the data shown in Jami et al., 2010), ORF name [associated with the P. chrysogenum Wisconsin 54-1255 genome (van den Berg et al., 2008)], Accession number, Description of the protein function, Score shown by Mascot database. In those spots where more than one protein was identified, these are shown separated by asterisks. Clicking over the spots, the protein information is shown in a new page, which allows to connect with the NCBI database.
This web page has been developed by Abel A. Cuadrado (University of Oviedo, Spain) supported by the data obtained from proteome analysis by Mohammad-Saeid Jami, Carlos García-Estrada, Carlos Barreiro, and Juan-F. Martín (Institute of Biotechnology of León and University of León, Spain).
Reference of the present web page:
Jami MS, García-Estrada C, Barreiro C, Cuadrado AA, Salehi-Najafabadi Z, Martín JF. The Penicillium chrysogenum extracellular proteome. Conversion
from a food-rotting strain to a versatile cell factory for white biotechnology. Mol Cell Proteomics. 2010 Dec;9(12):2729-44. Epub 2010 Sep 7.
Postal address of Jami MS, Barreiro C, García-Estrada C and Martín JF:
INBIOTEC (Institute of Biotechnology of Leon)
Parque Científico de León.
Avda. Real, 1
24006 León
Spain
e-mail: jf.martin@unileon.es
Postal address of Cuadrado AA:
Edificio Departamental 2 (Desp. 2.2.02)
Campus Universitario s/n
33204 Gijón
Asturias
Spain